
Single cell RNA sequencing
Multi-Organ Endothelial Cell Atlas Analysis and Liver-Specific Therapeutic Target Discovery
Executive Summary
This comprehensive analysis of the E-MTAB-8077 multi-organ endothelial cell dataset successfully identified and prioritized 118 liver-specific cell surface therapeutic targets through systematic integration of single-cell transcriptomics, spatial analysis, and druggability assessment. The study analyzed 38,983 endothelial cells from 10 organs, revealing distinct molecular specialization patterns and establishing a robust pipeline for liver-directed therapeutic interventions.
Dataset Characterization and Multi-Organ UMAP Analysis
Key Findings
Comprehensive atlas construction: Successfully integrated and analyzed 38,983 high-quality endothelial cells from 10 organs (brain, colon, heart, kidney, liver, lung, muscle, small intestine, spleen, testis) using 5,359 common genes, identifying 20 distinct clusters through graph-based clustering with resolution 0.5
Organ-specific clustering patterns: UMAP visualization revealed clear tissue-specific endothelial specialization with liver endothelial cells (4,168 cells, 10.7%) forming distinct clusters alongside organ-enriched populations from brain (Cluster 3), lung (Cluster 4), and liver (Cluster 5)
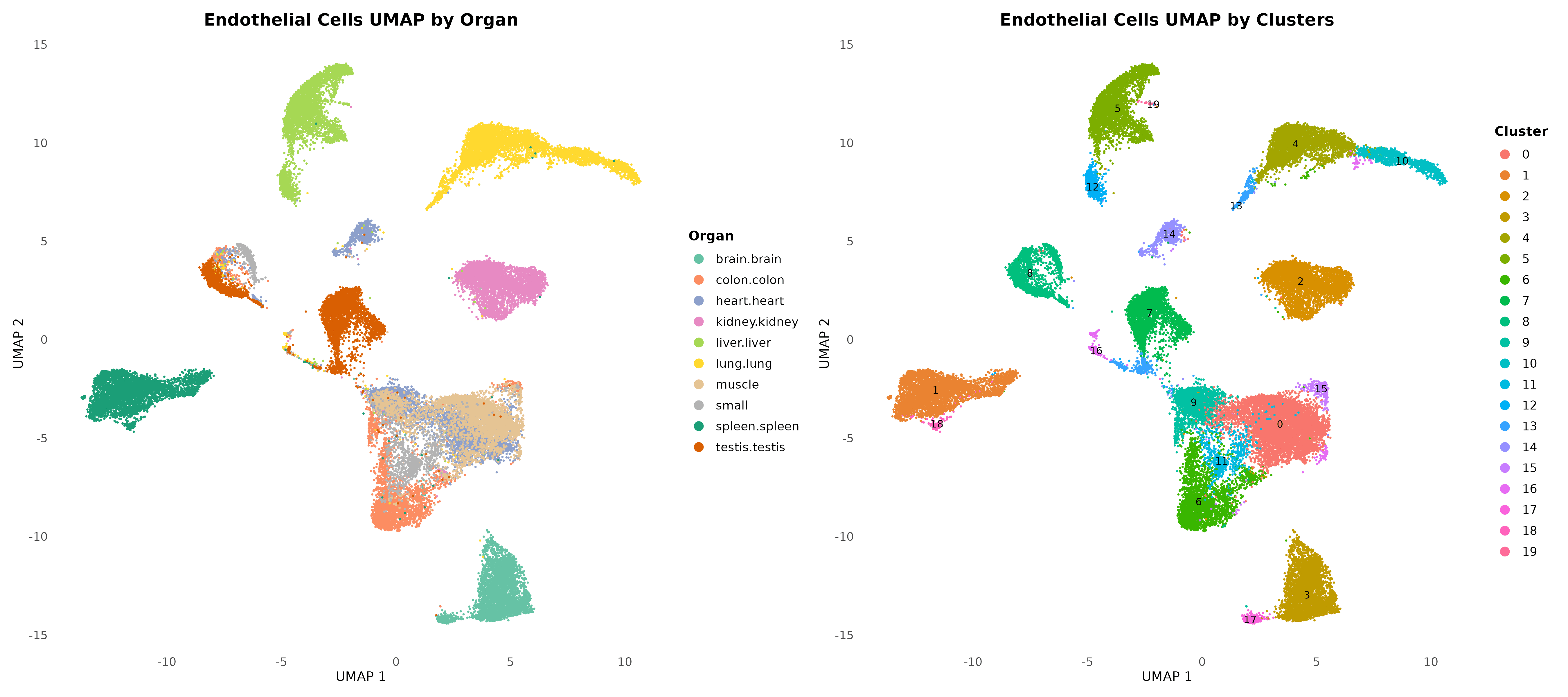
Dual organizational principles: Identified both highly organ-restricted clusters and pan-organ endothelial subtypes, indicating fundamental endothelial programs that transcend tissue boundaries while maintaining specialized functions for local microenvironments
Liver Endothelial Cell Molecular Specialization
Key Findings
Differential expression analysis: Identified 891 significantly upregulated genes in liver endothelial cells compared to other organs (p_adj < 0.05), with 482 high-confidence liver-specific genes showing LogFC > 0.5, demonstrating extensive molecular specialization
Top liver-specific markers: Lgmn (legumain) shows highest specificity with 3.68-fold enrichment (73.4% liver vs 13.8% others), followed by Apoe (3.37-fold), Lgals1 (3.18-fold), Cd59a (3.10-fold), and Man2a1 (2.91-fold), representing distinct functional categories
Functional pathway enrichment: Liver-specific genes cluster into lipid metabolism (Apoe, Agpat5), immune regulation (Lgmn, Cd59a, Il6st), lysosomal processing (Lgals1, Lamp2, Ctsl), and specialized transport (Slc29a1, Sepp1), reflecting hepatic physiological demands
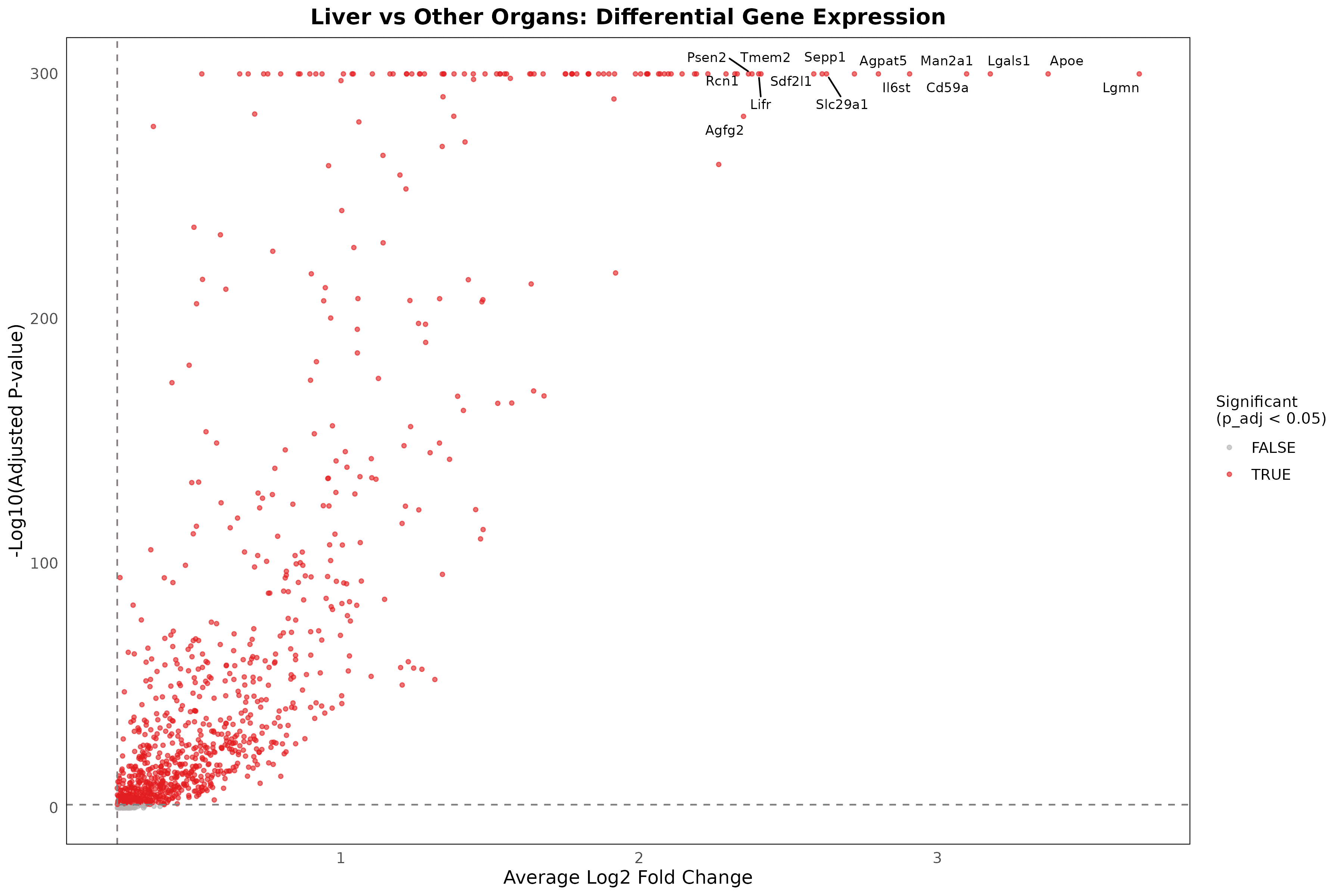
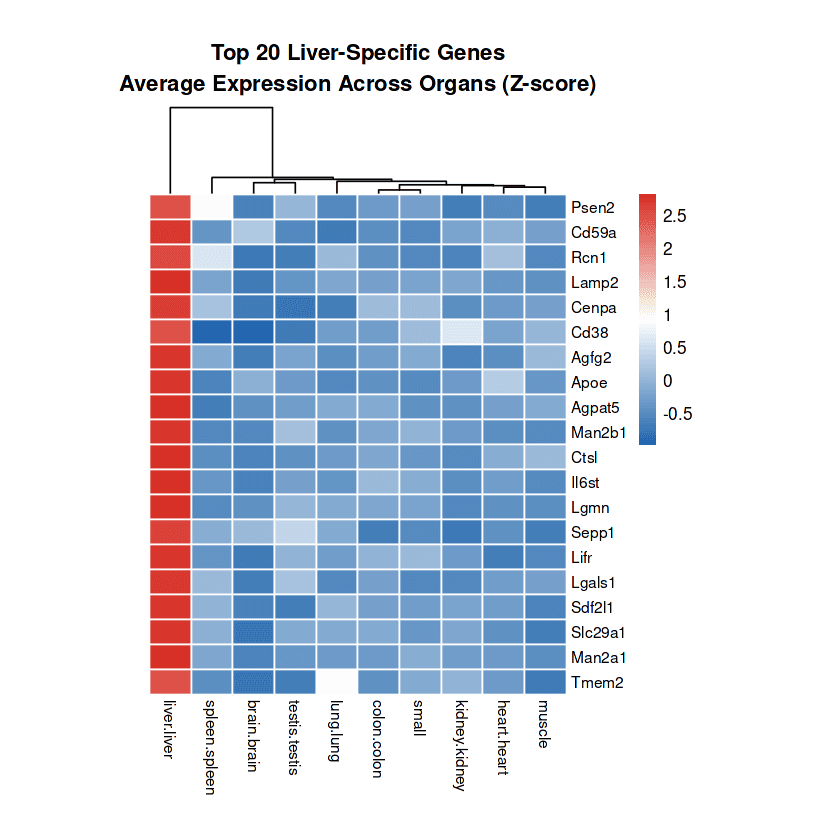
Cell Surface Therapeutic Target Discovery and Prioritization
Key Findings
Systematic target identification: Applied multi-database integration (GO terms, InterPro domains, biomaRt annotations) to identify 118 validated cell surface targets from liver-specific genes, establishing a comprehensive therapeutic pipeline
Therapeutic prioritization: Il6st (Interleukin-6 Signal Transducer) emerges as the top target with Priority score 3.00, combining high druggability (cytokine receptor), significant liver specificity (2.80-fold enrichment, 79.8% vs 25.8%), and established JAK/STAT pathway therapeutic precedent
Druggability stratification: Identified 6 high-druggability targets (Il6st, Lifr, Fcgrt, Flt4, Npr1, Ptprb) representing established receptor families, 5 medium-druggability emerging opportunities, and 107 novel targets spanning receptors (44%), transporters (15%), and membrane proteins (14%)
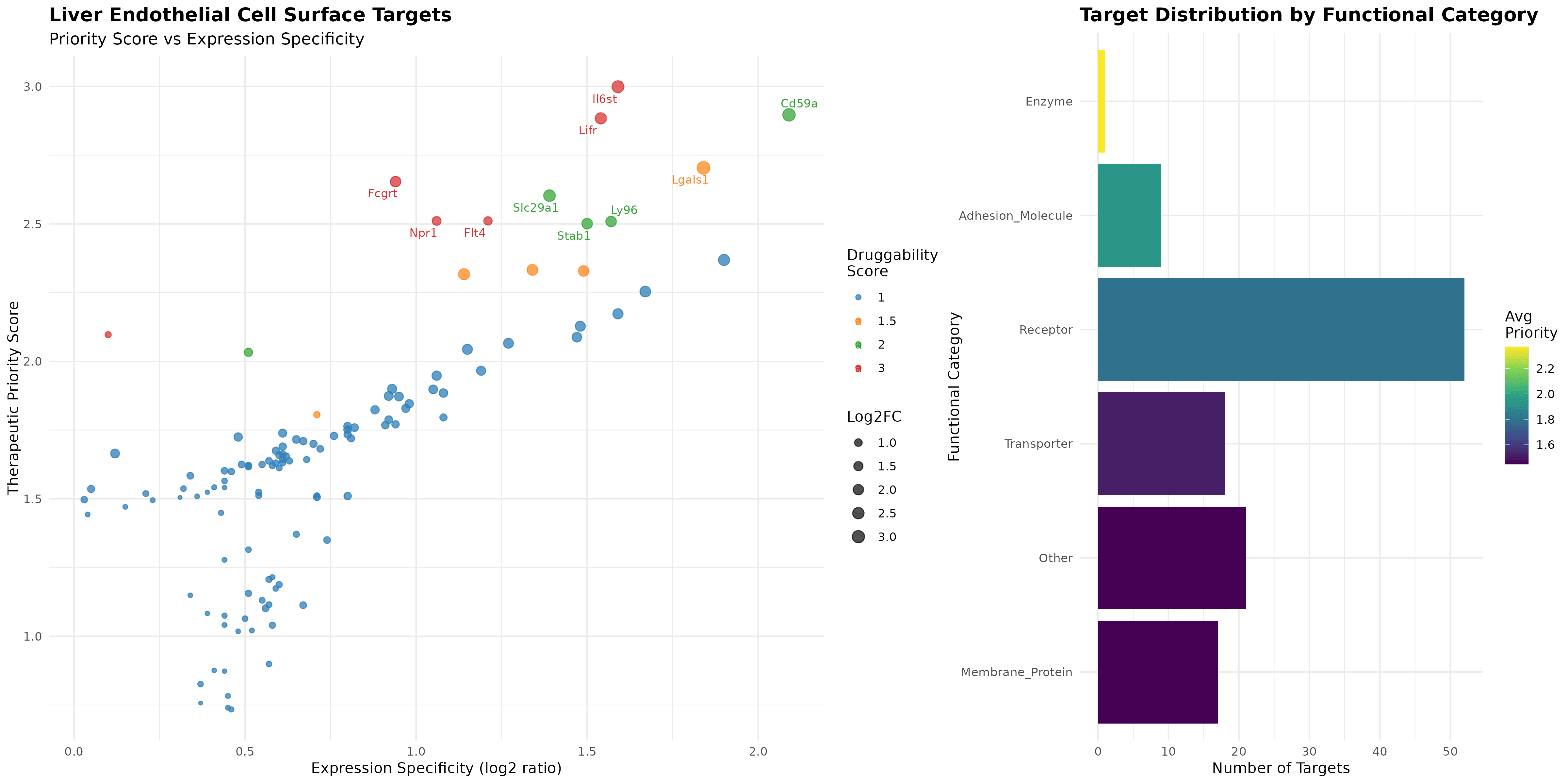
Spatial Expression Validation and Target Confirmation
Key Findings
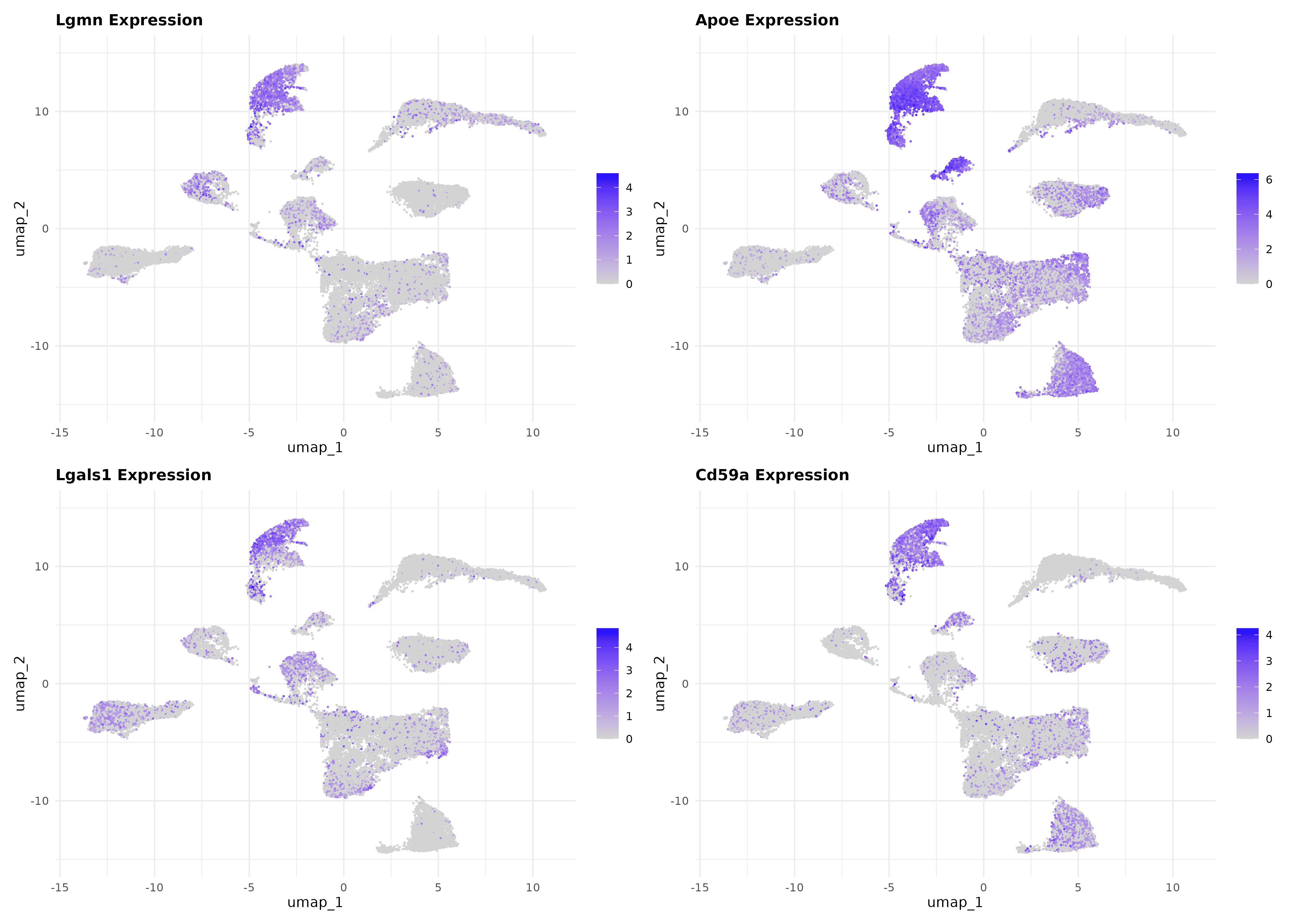
UMAP spatial validation: Successfully visualized 8 top-ranked liver endothelial cell surface targets on UMAP, confirming distinct liver-enriched expression patterns with clear spatial organization and minimal off-target distribution
Il6st optimal targeting confirmation: Top-priority target shows concentrated expression in liver-enriched UMAP clusters with sharp spatial boundaries, providing definitive single-cell validation of computational prioritization and supporting precision targeting strategies
Target hierarchy spatial validation: Top 4 targets (Il6st, Cd59a, Lifr, Lgals1) exhibit distinct UMAP localization patterns directly correlating with computational priority scores, ranging from highly liver-specific clusters to liver-enriched with moderate pan-organ expression
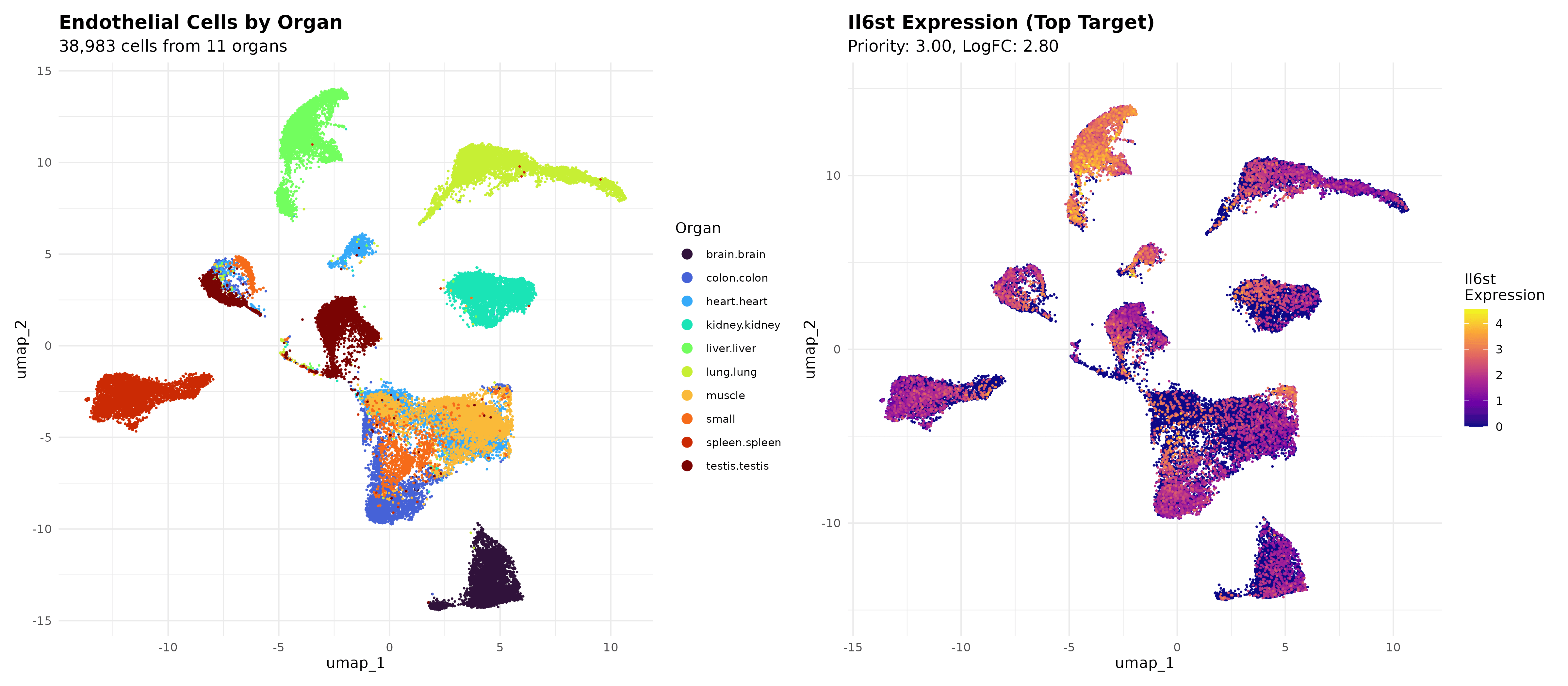
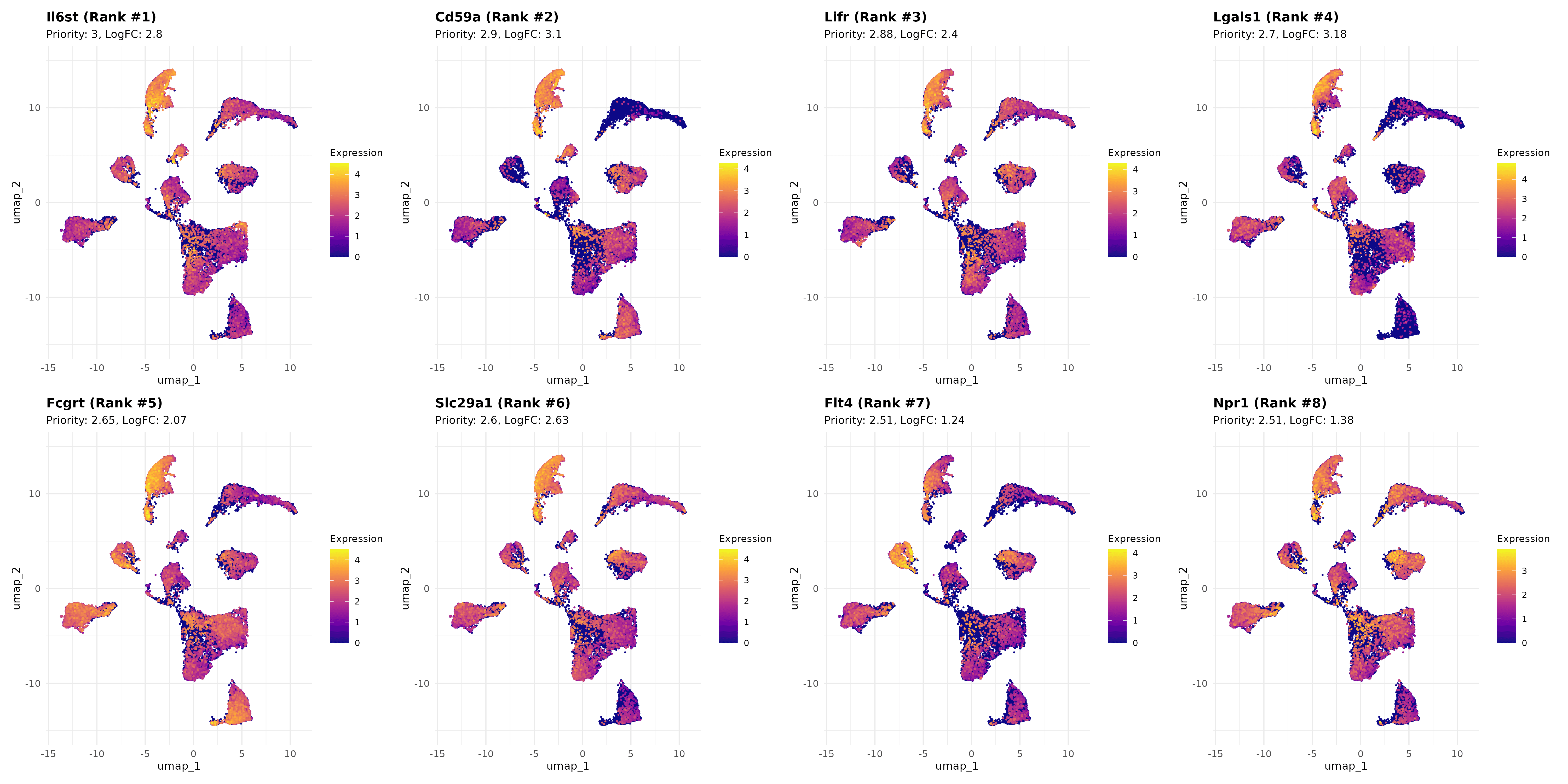
Interpretation
This comprehensive analysis establishes a robust framework for liver-directed therapeutic development through systematic identification of tissue-specific endothelial targets. Il6st represents the optimal immediate therapeutic opportunity with established JAK/STAT pathway druggability, exceptional liver specificity, and clear spatial validation, supporting anti-inflammatory interventions for liver diseases. The functional specialization hierarchy reveals liver endothelial cells as active signaling hubs with specialized programs for lipid metabolism, immune regulation, and transport functions rather than passive vascular barriers. Spatial expression validation through UMAP provides definitive confirmation that computational target prioritization accurately predicts tissue-specific therapeutic potential, with clear expression boundaries supporting precision medicine approaches.
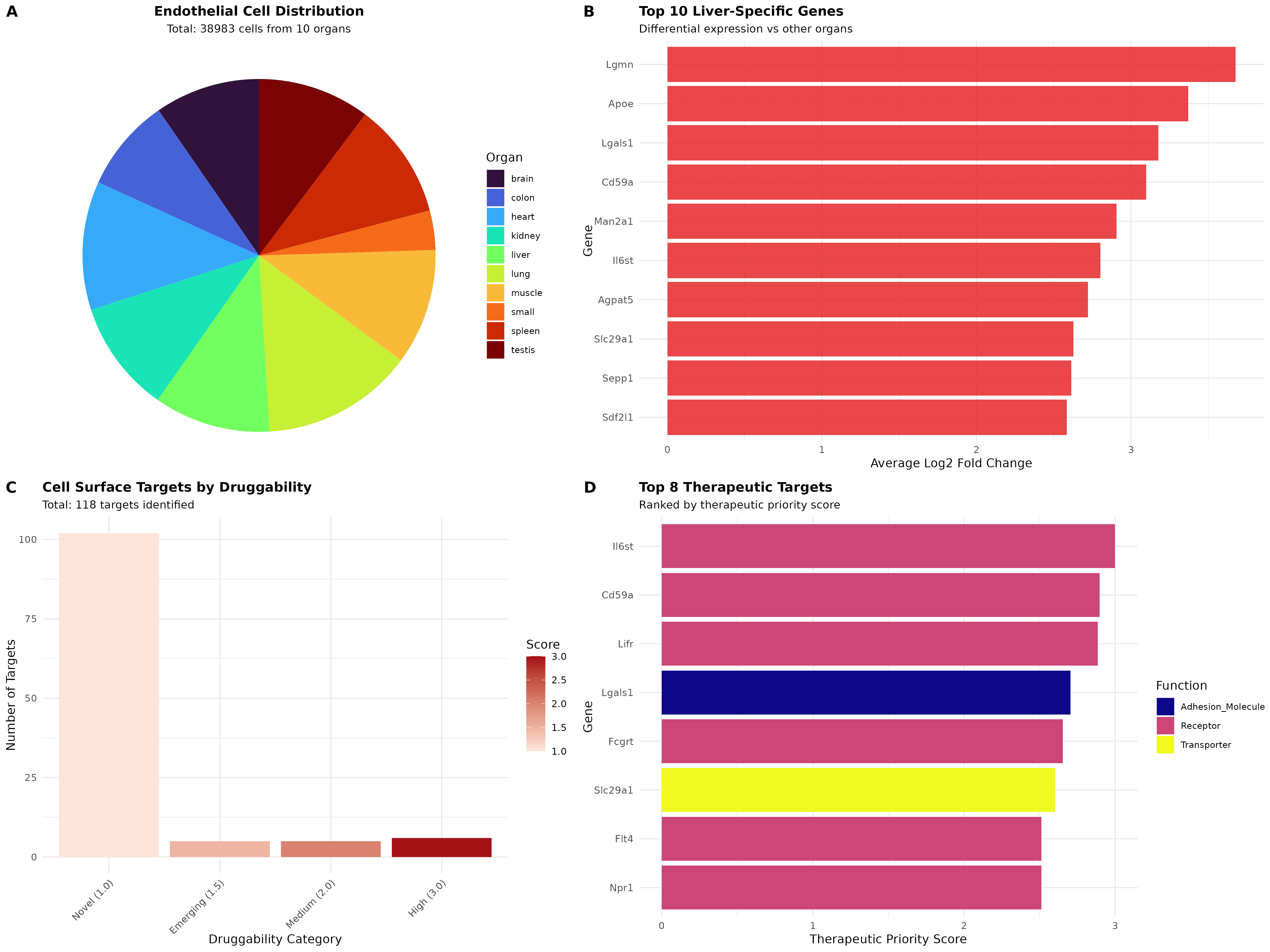
Strategic Therapeutic Implications and Clinical Translation
Immediate Development Opportunities
Phase I candidates: Il6st (JAK/STAT pathway) and Lifr (growth factor receptor) represent immediate therapeutic development opportunities with established pathway knowledge for inflammatory liver diseases and regenerative medicine applications
Precision targeting validation: Cd59a and Lgals1 demonstrate highest spatial specificity with minimal off-target expression, supporting improved therapeutic windows compared to systemic interventions for complement regulation and immune modulation
Combination therapy strategies: Multi-target approaches leveraging receptor + transporter combinations (52 receptors + 18 transporters) offer synergistic liver-directed therapeutic strategies for complex diseases including NASH, fibrosis, and autoimmune hepatitis
Recommended Next Steps
Functional validation studies: Prioritize Il6st, Cd59a, and Lifr for targeted perturbation experiments in liver endothelial cell models to confirm therapeutic efficacy and mechanism of action
Drug development pipeline: Initiate medicinal chemistry efforts for high-druggability targets while advancing preclinical validation of emerging opportunities (Lgals1, Igfbp4, Scarb2)
Clinical translation: Analyze target expression patterns in human liver disease samples to validate therapeutic relevance and establish biomarker potential for patient stratification
Intellectual property strategy: Develop comprehensive patent landscape analysis for top targets and liver-specific targeting approaches to support commercial development


